This is my first not-a-paper publication. I wrote it in a much more conversational style, which I greatly prefer.
After Randy Sheckman’s statements on journals such as Nature, Science, etc. I started wondering if there was a way to quantify what kinds of articles these journals publish. After all, the general perception (notwithstanding the allegations of inflated “brand value”) is that these journals publish high-quality science. I personally believe this is true, i.e., Nature and company do publish high-quality peer-reviewed science.
The trap that a lot of people fall into is believing that something not published in high-impact-factor (HIF) journals is of lower quality. I don’t believe this, so I wanted to test a different idea. My working hypothesis: that HIFs publish science that is of broad interest, so as to have a broad-enough readership to sustain their business model which relies on being very highly-cited (i.e. popular).
To test whether HIFs are “broad” we need two things: to define and measure “broadness” and to define “HIF”. We’ll start with the latter.
High Impact Factors
Some journals are simply widely known and almost universally recognized as “highly cited”. In medicine, those are the New England Journal of Medicine. Lancet. The British Medical Journal. JAMA. Nature. Science. Cell.
Household brand names, really, if you are in academic medicine. Their 2012 Impact Factors as computed by the Journal Citation Reports hover between 30 and 50. The Impact Factor, by the way, is the average number of citations an article published in the year n-1 and n-2 receives from articles published in year n. As an average, it’s subject to all kinds of problems. Suffice it to say that CA: A Cancer Journal for Clinicians, had a 2012 Impact Factor of 153… three times the “measly” 51 of the venerable New England Journal of Medicine. Such is the power of averages; a few very highly cited articles in a journal that publishes few articles, and you end up with a small journal with a higher IF than NEJM.
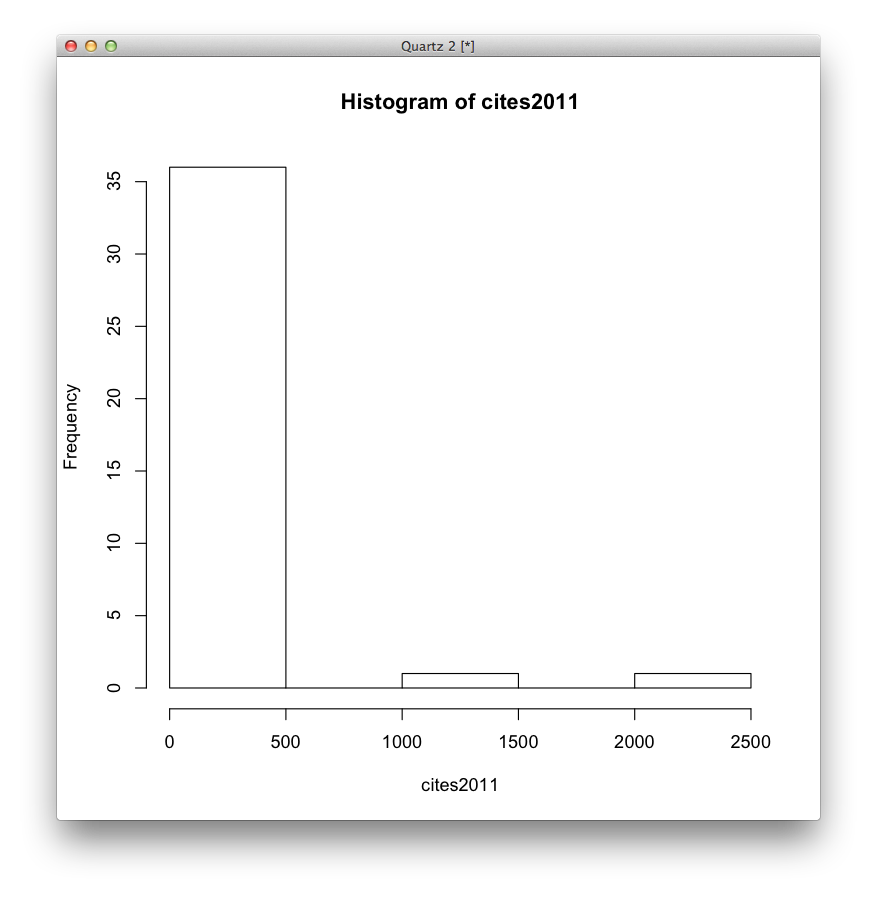
Here is the histogram of citations received, since publication (ergo, a number higher than the one used to compute the IF) for articles published in 2011 in CA: etc.
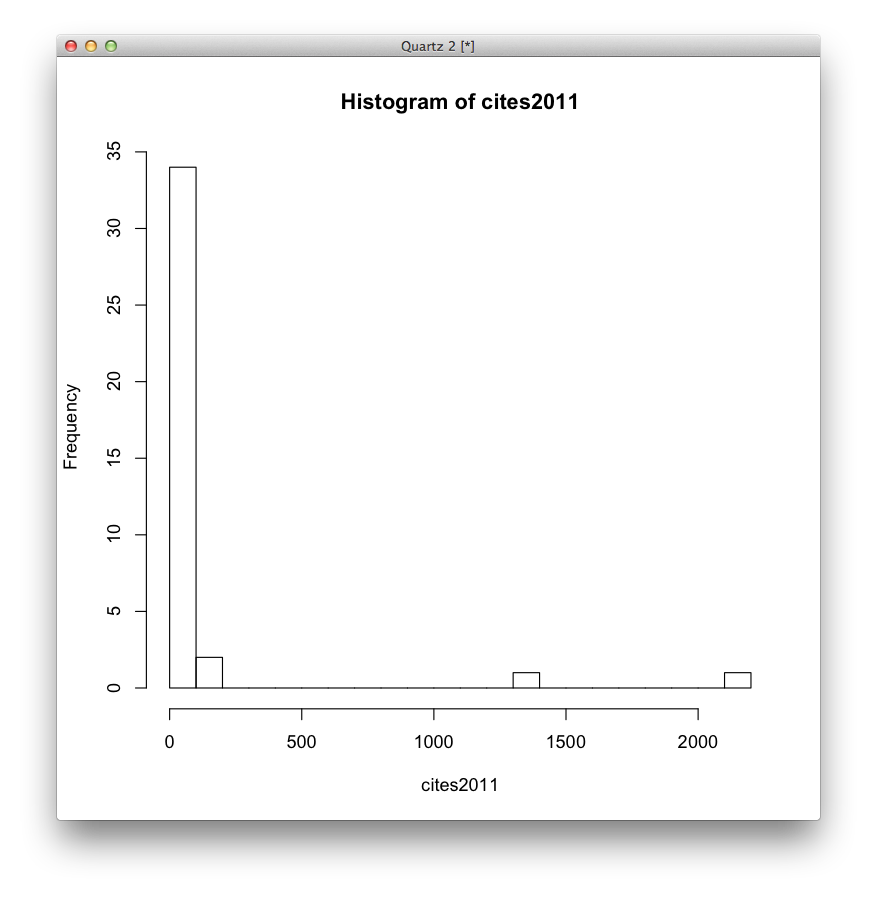
Fine, the first bar covers a pretty broad range that might justify the IF. Let’s try again, binning the data into 20 equally-spaced bins.
Convinced yet? The use of impact factors is madness, yet for some reason people’s careers depend on it. One article, on Global Cancer Statistics (very handy reference material) got 2000+ citations in 2012. Of course that will be highly cited, and it’s great, useful work. But it drags all the other 0-citation articles (I’m not claiming those are bad, mind you!) up into the HIF stratosphere.
Regardless, Impact Factors aren’t going anywhere any time soon. So let’s use a rule of thumb to define a HIF journal. Talk to any scientist at a large academic medical center – most will complain that the “best,” flagship journal in their field (however they choose to define it) has an IF of around 3. This is true for Biomedical Informatics, of course. On the other hand, commonly-known HIF journals have IFs of 30. I’ll use these two as a heuristic and declare that a Low Impact Factor (LIF) journal has an IF ≤ 4. A Medium Impact Factor (MIF) journal has an IF >4 and ≤ 10. And > 10 is a HIF journal.
Broadness
My working hypothesis here is that HIFs cover topics that are of broader interest than LIFs. Under this hypothesis, LIFs are highly technical, thus having smaller readerships who -in turn- have small readerships, getting less citations.
So how general is the topic of a scientific article? MEDLINE can help. Every article in MEDLINE is tagged with MeSH headings: concepts from the Medical Subject Headings vocabulary assigned by a professional, highly-trained medical librarian describing what topics are covered in an article. There’s actually two kinds of MeSH headings for a MEDLINE article: Major Topics and non-Major Topics. Major Topics are what an article is about; non-Major topics are important things mentioned in an article.
<MeshHeadingList>
<MeshHeading>
<DescriptorName MajorTopicYN="N">Algorithms</DescriptorName>
</MeshHeading>
<MeshHeading>
<DescriptorName MajorTopicYN="N">Information Storage and Retrieval</DescriptorName>
<QualifierName MajorTopicYN="Y">statistics & numerical data</QualifierName>
</MeshHeading>
<MeshHeading>
<DescriptorName MajorTopicYN="N">Internet</DescriptorName>
</MeshHeading>
<MeshHeading>
<DescriptorName MajorTopicYN="N">Medical Subject Headings</DescriptorName>
<QualifierName MajorTopicYN="N">utilization</QualifierName>
</MeshHeading>
<MeshHeading>
<DescriptorName MajorTopicYN="N">PubMed</DescriptorName>
<QualifierName MajorTopicYN="Y">statistics & numerical data</QualifierName>
</MeshHeading>
</MeshHeadingList>
This is an example from the XML output of the record for one of my own articles. The nice thing about MeSH is that it is a thesaurus (in the Information Retrieval sense) – things have parents. For example, the MeSH term for Medical Subject Headings themselves is classified as
(you can look these up in the MeSH Browser). The relationships are IS_A, which means that Medical Subject Headings IS_A kind of Subject Headings, which IS_A kind of Vocabulary, Controlled, which IS_A kind of Documentation, which in turn IS_A kind of Information Service which itself IS_A Information Science. You may or may not agree with the exact arrangement in this taxonomy; for my purposes, I just want you to note that Medical Subject Headings is a very detailed concept: it’s five levels under Information Science in the MeSH tree.
You also need to know that the MeSH indexers are required to use the most detailed term they can. This means that the article didn’t just mention any Subject Headings: it mentioned something more specific. So an article like mine above was more specific than one that just dealt with Subject Headings, because it dealt with Medical Subject Headings.
You can probably see where this is going. We can use the depth of the terms assigned to an article as a measure of its generality or lack thereof – of its “broadness.”
MeSH terms can be in multiple trees at once. In order to simplify things a little, we’ll choose the deepest term position possible, effectively assuming that all concepts are at their most specific. In other words, when an article is tagged with the MeSH concept “eye” we’ll assume Body Regions->Head->Face->Eye for a depth of 3 (counting from 0) instead of Sense Organs->Eye for a depth of 1. This is arbitrary, but if we can still find a difference in depth despite assuming that everything is as specific as possible, it should be a robust result.
For our purposes, we’ll focus on Descriptors, which are the actual unmodified topics, and we’ll only take Major topics – things articles are about. We’ll collect them for all articles in every journal.
Methods
You’ll need a copy of PubMed/MEDLINE in XML format for this one. Don’t worry, it’s “just” 13 GB. Hard drives are cheap. You’ll also need a computer with 8 GB of RAM at a minimum, unless you want to edit my code and make it more efficient. It’s probably easier to just buy more RAM. You know you want to, anyway.
You’ll then need to install xmltodict. How to install Python packages is beyond the scope of this article, but if
pip install xmltodict
or
easy_install xmltodict
don’t help you, you’ll have to do some Googling. Then you’ll want to take MEDLINE and turn it into a Journal name->MeSH Term->count dictionary. Who wouldn’t? Grab a copy of read_medline.py and build_journal_term_database.py. Put them on the aforementioned computer with lots of RAM (and many cores help too, by the way), and run
python build_journal_term_database.py <PATH_TO_MEDLINE_BASELINE_HERE>
You can use pypy instead of python for a speed boost, at the cost of Even More Memory. When the process ends, there’ll be a file called journal_major_mesh_terms.pickle in the same directory. It’s only 131 MB on my machine.
In part 2, we’ll compute some statistics on this dictionary.